QuantArray®, an advanced qPCR method, quantifies a broad spectrum of contaminant degrading microorganisms and functional genes in a single analysis for more comprehensive and cost-effective evaluation of biodegradation potential. Microbial Insights offers four versions of QuantArray® for the Environmental Remediation industry:
QuantArray® -Petro
QuantArray® -Petro quantifies a suite of functional genes involved in aerobic and anaerobic biodegradation of BTEX, PAHs, and other petroleum hydrocarbons.
QuantArray®-Chlor
QuantArray® -Chlor quantifies key microorganisms (e.g., Dehalococcoides, Dehalobacter, etc.) and functional genes (e.g., vinyl chloride reductase, methane monooxygenase, etc.) to assess potential for reductive dechlorination and aerobic cometabolism of chlorinated solvents such as TCE.
QuantArray® -BGC
QuantArray® -BGC quantifies – simultaneously in a single analysis – a broad spectrum of microorganisms and genes involved in biogeochemical processes.
QuantArray® -NSZD
QuantArray® -NSZD quantifies – simultaneously in a single analysis – a broad spectrum of microorganisms, processes and genes involved in biodegrading LNAPL for assessing NSZD.

QuantArray® ADVANTAGES:

ACCURATE
Direct analysis of sample DNA removes the need to grow the bacteria, thus eliminating biases associated with traditional approaches (e.g., plate counts and MPNs).
QUANTITATIVE
Absolute quantification of the concentrations of specific microorganisms and functional genes encoding enzymes responsible for contaminant biodegradation gives site managers a direct line of evidence to evaluate remediation options and monitor remedy performance. Results reported as cells/mL, cells/g, etc.

COST EFFECTIVE
QuantArray®-Chlor and QuantArray®-Petro simultaneously quantify suites of key microorganisms and functional genes responsible for biodegradation of chlorinated solvents and petroleum hydrocarbons, respectively. QuantArray ® saves money because site managers can make highly informed decisions based on comprehensive assessment of anaerobic and aerobic biodegradation of a variety of contaminants (e.g., PCE, TCE, DCE, VC, TCA, DCA, CF, etc.) by a multitude of microorganisms (e.g., Dehalococcoides, Dehalobacter, Dehalogenimonas, etc.) and pathways in a single analysis.

INFORMATIVE
Is that a low, medium or high concentration of contaminant degraders? With the MI Database, clients can retrieve percentile rankings of their QuantArray® results to answer that question based on the tens of thousands of samples MI has received from sites around the world.

SENSITIVE
Practical Detection Limits (PDL) are as low as 100 cells per sample with a dynamic range over seven orders of magnitude. Low detection limits are particularly important when evaluating whether bioaugmentation is needed or an unnecessary expense.

SPECIFIC
Target specific bacterial groups (e.g., Dehalococcoides, Dehalobacter, Dehalogenimonas) and functional genes (e.g., vinyl chloride reductases, anaerobic benzene carboxylase) responsible for contaminant biodegradation.

FLEXIBLE
Analysis can be performed on almost any type of sample (water, soil, sediments, Bio-Traps®, and others).
HOW TO USE QuantArray®:
Along with contaminant concentrations and geochemical parameters, the concentrations of specific microorganisms and functional genes responsible for contaminant biodegradation is a key component of remedy selection and performance monitoring at sites impacted by chlorinated solvents or petroleum hydrocarbons.
Use QuantArray® to help answer…
- How feasible is MNA? Is enhanced bioremediation needed? Is bioaugmentation needed?
- What are the concentrations of contaminant degraders under existing conditions?
- Are degrader concentrations greater in impacted wells than background wells?
- Based on the MI Database, are concentrations of contaminant degraders low, medium, or high in the plume?
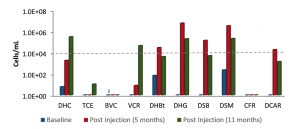
- Is enhanced bioremediation effective?
- Did concentrations of contaminant degraders and functional genes increase in response to treatment?
- Are additional types of degraders and functional genes now detected that were previously below detection limits?